- ⌂
-
Le DPM
Les Equipements
L'Environnement
Zoom sur... Le Bâtiment André Rassat
Nommé d'après une figure tutélaire de la chimie grenobloise, ce bâtiment est recouvert d’une double peau en feuille métallique qui apporte une protection thermique sur 3 côtés et crée une unité architecturale favorisant l'intégration parmi les arbres du site.
-
Thématiques
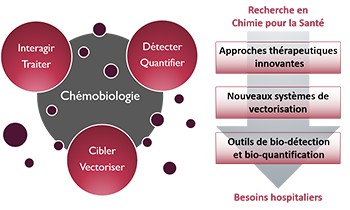
Le Thème
« Approches thérapeutiques innovantes »De nouvelles solutions thérapeutiques, de la cible biomacromolécu-laire émergente aux approches originales pour traiter les maladies
Le Thème
« Nouveaux systèmes de vectorisation »Combiner les propriétés d’inclusion de principes actifs, de franchisse-ment de barrières, d’adressage et de relar-gage en milieu vivant
Le Thème
« Outils de bio-détection et bio-quantification »Des dispositifs analytiques originaux pour la détection de cibles, de l’ion au micro-organisme en milieu complexe
Zoom sur... La Chémobiologie
-
Équipes
L'Équipe « COMET »
« COMET » développe la conception rationnelle, la synthèse et/ou l'extraction de composés à forte diversité/complexité comme nouveaux agents thérapeutiques et outils moléculaires pour la pénétration cellulaire ou la détection de biomolécules, actifs in vivo.L'Équipe « NOVA »
« NOVA » utilise des acides nucléiques fonctionnels comme éléments de reconnaissance pour des applications thérapeutiques ou diagnostiques, comme la sélection d'oligonucléotides, ou le développement de dispositifs d'analyses et de nanovecteurs.Les Services
-
Productions
Les Publications
La Vulgarisation
Les JSM
Zoom sur... La 12ème JSM (15 juin 2023)
Le DPM organise des journées scientifiques consacrées au médicament. L'objectif est de rassembler les spécialistes académiques et industriels autour d'une thématique. 2023 : Apports de la Chimie Click et de la Lumière en Chemobiologie
-
Partenariats
Les Formations
Les Consortiums
Les Financements
Zoom sur... L'environnement Grenoblois
Le DPM est un acteur central sur le bassin grenoblois en chimie, biologie et santé, lié au CHU Grenoble Alpes et à de nombreuses autres organisations : Pole de Recherche CBS, ICMG, Labex ARCANE, EUR CBH, Institut Carnot Polynat, Réseau GREEN.
Article
- Projet
- Ahcène BOUMENDJEL, Romain HAUDECOEUR, Marine PEUCHMAUR,
- Titre
- Structure-Activity Relationships in the Development of Allosteric Hepatitis C Virus RNA-Dependent RNA Polymerase Inhibitors: Ten Years of Research
-
[Full paper
 ]
] - Auteurs
- R. Haudecoeur, M. Peuchmaur, A. Ahmed-Belkacem, J.-M. Pawlotsky,A. Boumendjel
- Edition
- Med. Res. Rev. 2013, 33, 934-984.
- Année
- 2013
- Résumé
- A review. Hepatitis C is a viral liver infection considered as the major cause of cirrhosis and hepatocellular carcinoma (HCC). Hepatitis C virus (HCV) possesses a single pos. strand RNA genome encoding a polyprotein composed of approximatively 3000 amino acids. The polyprotein is cleaved at multiple sites by cellular and viral proteases to liberate structural and nonstructural (NS) proteins. NS5B, the RNA-dependent RNA polymerase (RdRp), which catalyzes the HCV RNA replication has emerged as an attractive target for the development of specifically targeted antiviral therapy for HCV (DAA, for direct-acting antivirals). In the last 10 years, a growing no. of non-nucleoside compds. have been reported as RdRp inhibitors and few are undergoing clin. trials. Over the past 5 years, several reviews were published all describing potentially active mols. To the best of our knowledge, only one review covers the structure-activity relationships.1 In this review, we will discuss the reported non-nucleoside mols. acting as RdRp inhibitors according to their chem. class esp. focusing on structure-activity relationship aspects among each class of compds. Thereafter, we will attempt to address the global structural requirements needed for the design of specific inhibitors of RdRp.




 Annuaire
Annuaire Contact
Contact Plan d'accès
Plan d'accès ENG
ENG Login
Login



